The function of cap structure and capping efficiency detection
The structure of mRNA mainly includes the 5' cap , the 5' untranslated region (UTR), and the open reading frame (ORF), 3' end untranslated region and Poly(A) tail. All mature eukaryotic mRNA have a methylguanosine cap (m7G) structure at the 5' end. Capping can help mRNA be smoothly transported out of the nucleus, increase mRNA stability, prevent it from being degraded by ribonuclease, and is crucial for protein expression.

At present, the main method used is to first synthesize a probe containing biotin that matches the first 50nt of the 5'UTR region, let the probe bind to mRNA, then perform enzyme cleavage with RNase H, enrich the probe with the 5’ cleavage products using streptavidin magnetic beads specifically, and finally eluted in water . The purified 5’ cleavage products are visualized using mass spectrometry(LC-MS).

The role of Poly(A) structure and Poly(A) distribution detection principle
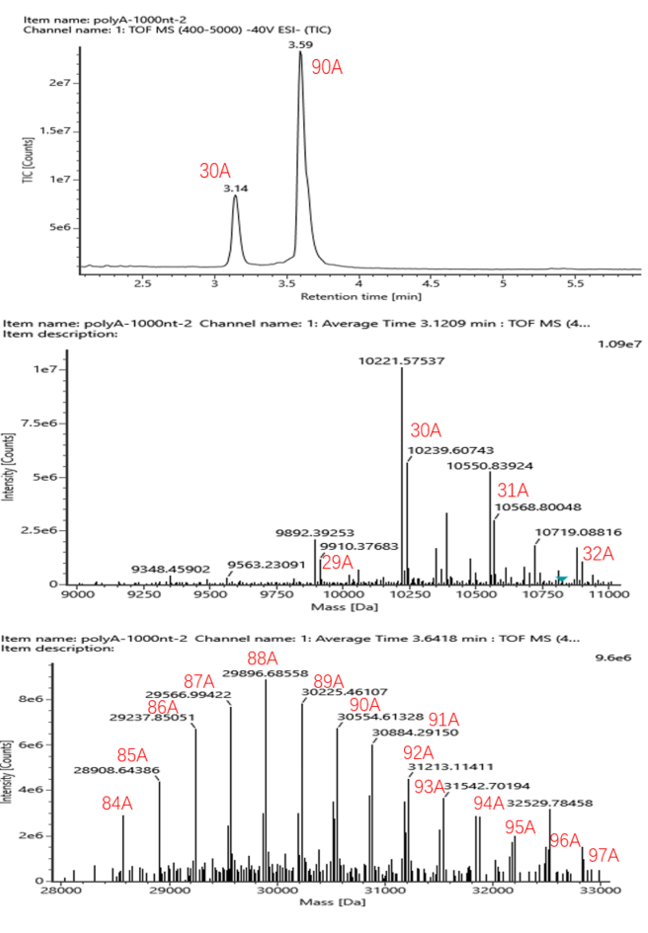
Eukaryotic mRNAs often terminate with a stretch of adenosines on their 3′ end which can vary in length from 20 to over 200 bases depending on the species.
It has been postulated that longer tails increase translation efficiency, provide mRNA stability, and can possibly affect immune response although there has been some recent research suggesting that tail length of mRNA and translation efficiency are linked only during specific cell development stages。
At present, RNase T1 is mainly used for enzyme digestion, and the polyA fragments after enzyme digestion are enriched with Oligo DT. After elution, the polyA fragments enriched on the filler are entered into LC-MS for analysis.
Third-party testing services
Quality control is one of the most important aspects of drug development,Hzymes provides third-party quality control services for mRNA drugs.
■ Self-established quality control analysis platform ■ mRNA capping efficiency detection ■ mRNA Poly(A) distribution detection
■ Plasmid analysis method development and validation ■ Development and validation of analytical methods for LNP component content analysis
■ Product and process impurity studies ■ Raw and auxiliary materials testing ■ One-stop CMC solution




Service items | Periodicity(Working days) | Deliverables | Criteria |
Probe Design & synthesis | 5 | Probe | Determined by customer requirement |
Capping efficiency Detection | 3-10 | Detection report | |
| Poly(A) distribution detection | 3-10 | Detection report |
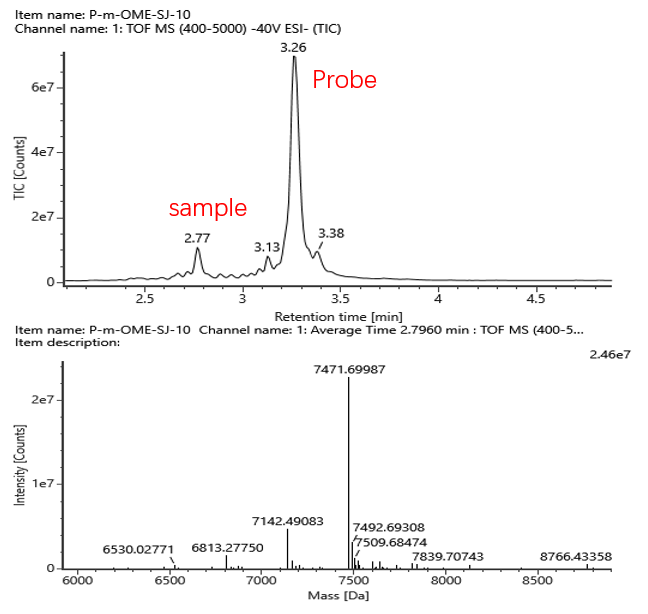
Name | Response | Observed mass (Da) | Expected mass (Da) | Mass error (mDa) | Mass error (ppm) | %Cap1 |
Monphos | 38908 | 7004.385 | 7004.297 | 88.1 | 12.6 | 95.1% |
Triphos | 1052878 | 7164.479 | 7164.256 | 223.1 | 31.1 | |
Triphos_a | 506256 | 6835.276 | 6835.05 | 225.3 | 33.0 | |
Diphos_a | 101498 | 6755.327 | 6755.071 | 256.5 | 38.0 | |
cap1 | 23880426 | 7471.694 | 7471.551 | 143.2 | 19.2 | |
cap1_gaa | 1924043 | 6468.059 | 6467.933 | 125.6 | 19.4 | |
cap1_a | 5077712 | 7142.483 | 7142.345 | 138.5 | 19.4 | |
cap1_aa | 2301145 | 6813.278 | 6813.139 | 139.1 | 20.4 |


